In response to stress, mammalian cells make one of three fundamental decisions – die, survive or thrive. Critical determinants of these seemingly distinct cell fates include the induction of signaling cascades and selective expression of perturbation-responsive genes, which together help restore ribostasis, proteostasis and cellular homeostasis. However, the mechanisms by which cells activate these processes and how cells selectively regulate gene expression to make survival decisions remains unclear. In addition, emerging evidence suggests that the dynamic reorganization of functionally nebulous, RNA- and protein-bearing, membrane-less compartments is an evolutionarily conserved hallmark of stress. Alterations in any of these stress response mechanisms are directly linked to aging and various (neurodegenerative and neoplastic) diseases. Elucidating the molecular bases of these aberrations and pathobiology will pave the way for new and effective diagnostics and therapeutics. Our lab seeks to understand how mammalian cells of various tissue contexts maintain homeostasis and how stress response programs influence cellular decisions in physiology and pathology.
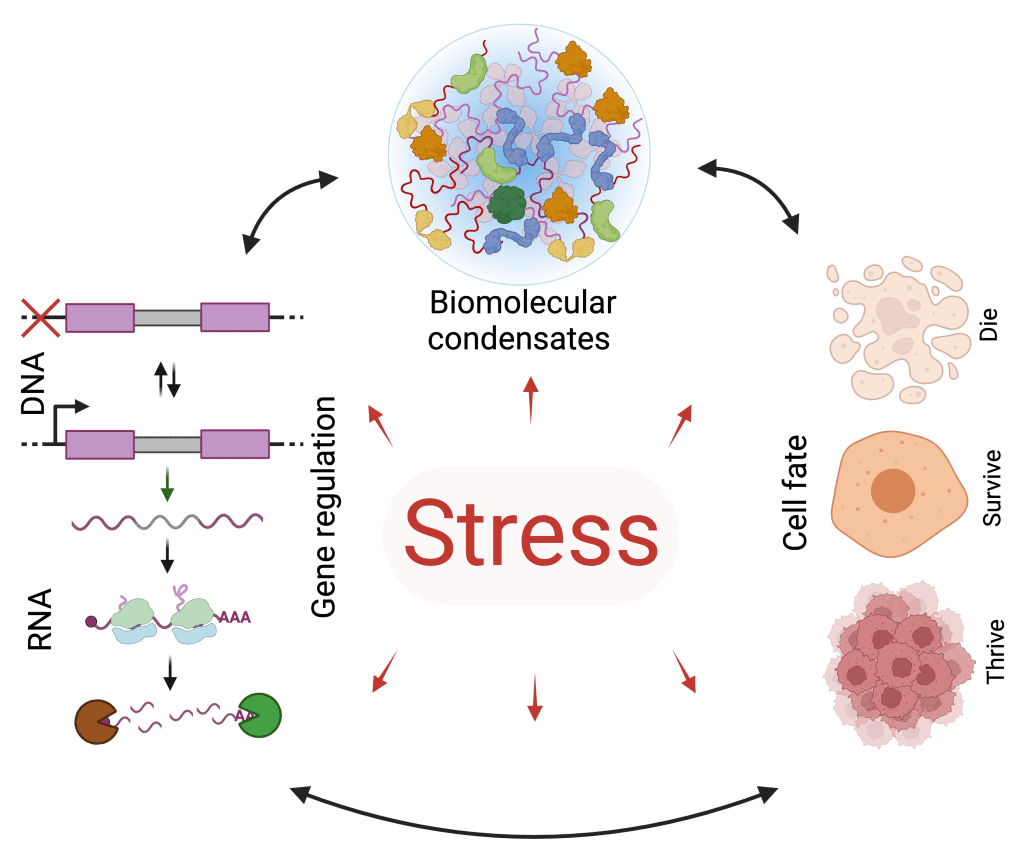
Our research seeks to address the following questions:
- How do cells achieve transcriptional and translational selectivity during and after stress?
- How do stress-induced biomolecular condensates form and function?
- Why do some cells die, while others survive or seemingly thrive after stress?
- How are stress response programs impaired during aging and degenerative diseases?
- How are stress response mechanisms misappropriated in cancer?
- How do condensates contribute to physiology and disease?
- Can stress response programs and condensates be targeted for diagnostics and therapeutics?
To address these pressing questions, we use an interdisciplinary approach that lies at the cusp of physical and life sciences. Herein, we utilize in vitro, cultured cell, animal and human tissue models for our research. We probe these models with an arsenal of contemporary imaging- and omics-based technologies that we develop and deploy, and combine them with classical biochemical and biophysical tools. Notably, we use single-molecule imaging, super-resolution microscopy, high-content screening, single-cell sequencing and spatial omics among other cutting-edge technologies to dissect the spatiotemporal dynamics of stress response. The ultimate aim is to leverage our foundational and translational research for diagnostic and therapeutic applications.
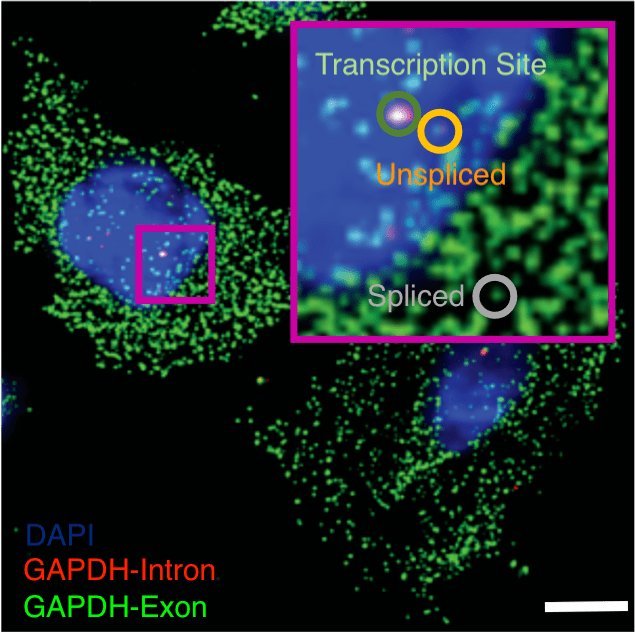
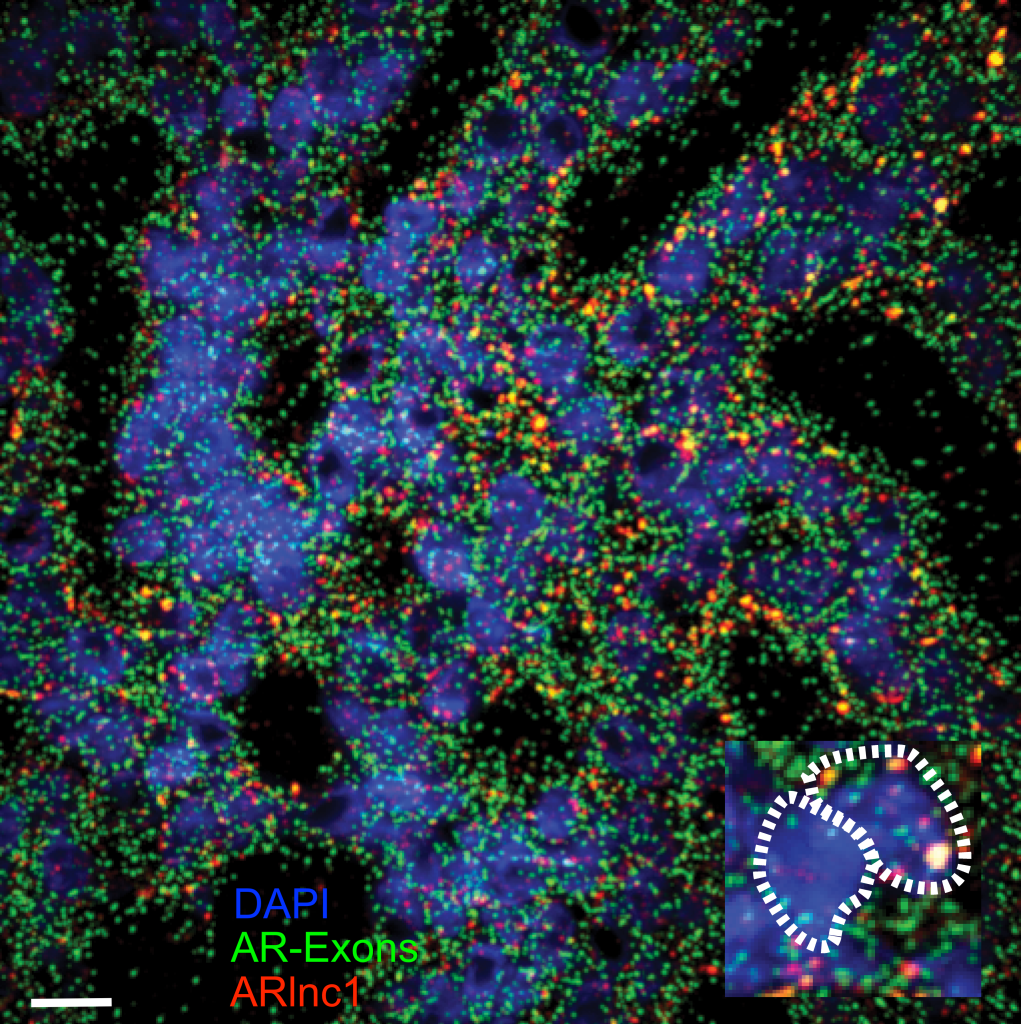
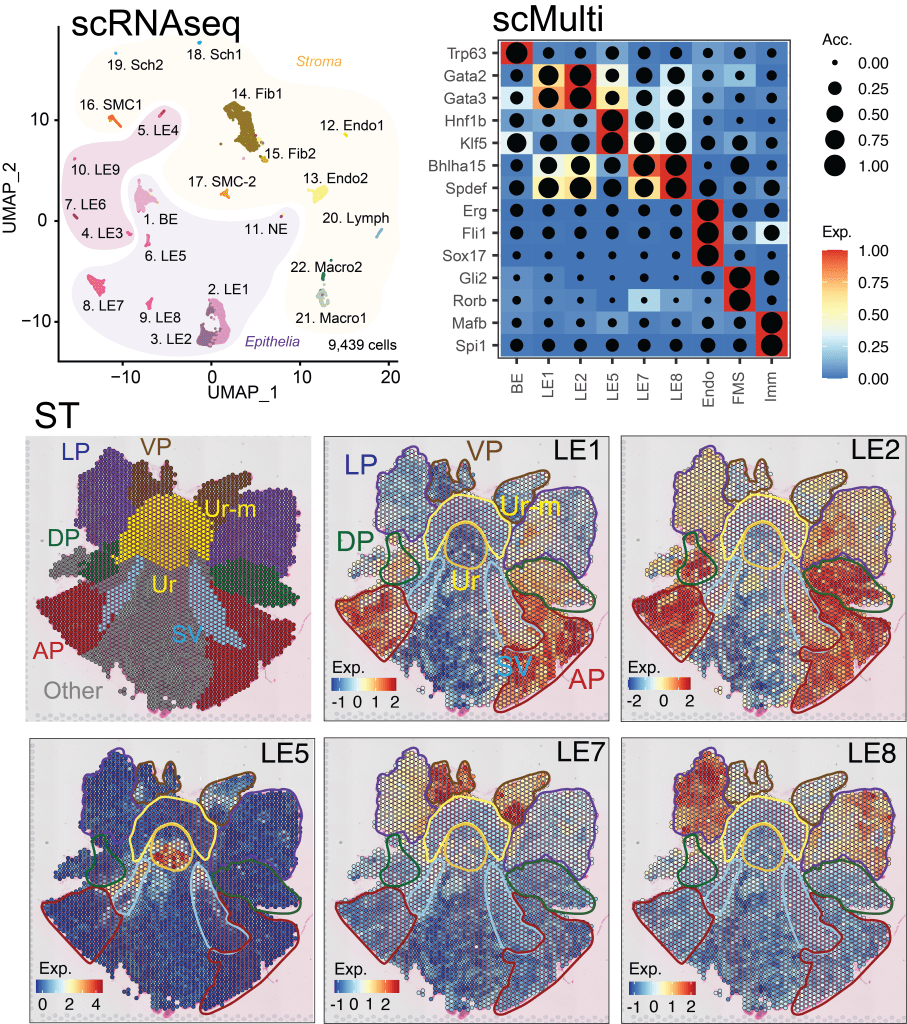
Interrogating gene regulation across spatial and biological scales. (Left – Right) Nanoscale imaging of GAPDH mRNA metabolism in U2-OS cells with single-molecule fluorescence in situ hybridization, scale 10 μm. Nanoscale imaging of the androgen receptor (AR) mRNA and AR-regulated long non-coding RNA (ARlnc1) in patient-derived, castration resistant prostate cancer sections using single-molecule fluorescence in situ hybridization, scale 30 μm. Single cell RNA sequencing (scRNAseq), single cell multiomics (scMulti) and spatial transcriptomics (ST) of the mouse prostate.
Measuring subcellular dynamics. (Left – Right) Mesoscale time-lapsed imaging (100 nm spatial resolution over 1 h) of subnuclear condensates. Single molecule tracking (30 nm spatial resolution and 30 ms temporal resolution) of a transcription factor (HSF1) in the nucleoplasm and at condensates. Mesoscale imaging (100 nm spatial resolution and 100 ms temporal resolution) of GFP-DCP1A (mRNA decapping coactivator) undergoing hyperosmotic phase separation and ensuing rescue under isosmotic conditions, within living U2-OS cells.